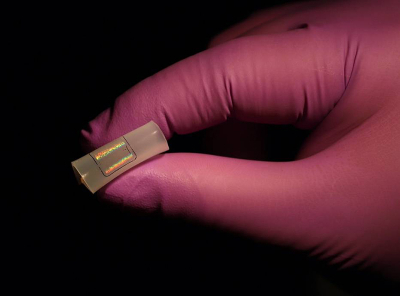
A COVID-19 sensor developed at Johns Hopkins University could revolutionise virus testing by adding accuracy and speed to a process that frustrated many during the pandemic. The sensor, which requires no sample preparation and minimal operator expertise, offers a strong advantage over existing testing methods, especially for population-wide testing.
“The technique is as simple as putting a drop of saliva on our device and getting a negative or a positive result”, said Ishan Barman, from Johns Hopkins University. “The key novelty is that this is a label-free technique, which means no additional chemical modifications like molecular labelling or antibody functionalisation are required. This means the sensor could eventually be used in wearable devices.”
Barman says the new technology, which is not yet available on the market, addresses the limitations of the two most widely used types of COVID-19 tests: PCR and rapid tests. PCR tests are highly accurate, but require complicated sample preparation, with results taking hours or even days to process in a laboratory. On the other hand, rapid tests, which look for the existence of antigens, are less successful at detecting early infections and asymptomatic cases and can lead to erroneous results.
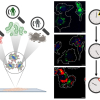
The sensor is nearly as sensitive as a PCR test and as convenient as a rapid antigen test. During initial testing, the sensor demonstrated 92 % accuracy at detecting SARS-COV-2 in saliva samples—comparable to that of PCR tests. The sensor was also highly successful at rapidly determining the presence of other viruses, including H1N1 and Zika. The sensor is based on large area nanoimprint lithography, surface enhanced Raman spectroscopy (SERS) and machine learning. It can be used for mass testing in disposable chip formats or on rigid or flexible surfaces.
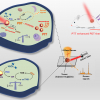
Key to the method is the large-area, flexible field enhancing metal insulator antenna (FEMIA) array developed by the group. The saliva sample is placed on the material and analysed using SERS. Because the nanostructured FEMIA strengthens the virus’s Raman signal significantly, the system can rapidly detect the presence of a virus, even if only small traces exist in the sample. Another major innovation of the system is the use of advanced machine learning algorithms to detect very subtle signatures in the spectroscopic data that allow researchers to pinpoint the presence and concentration of the virus.
“Label-free optical detection, combined with machine learning, allows us to have a single platform that can test for a wide range of viruses with enhanced sensitivity and selectivity, with a very fast turnaround”, said Debadrita Paria from Johns Hopkins University.
The sensor material can be placed on any type of surface, from doorknobs and building entrances to masks and textiles.
“Using state of the art nanoimprint fabrication and transfer printing we have realised highly precise, tuneable and scalable nanomanufacturing of both rigid and flexible COVID sensor substrates, which is important for future implementation not just on chip-based biosensors but also wearables”, said David Gracias, a professor of chemical and biomolecular engineering from Johns Hopkins. He says the sensor could potentially be integrated with a hand-held testing device for fast screenings at crowded places like airports or stadiums.
“Our platform goes beyond the current COVID-19 pandemic”, said Barman. “We can use this for broad testing against different viruses, for instance, to differentiate between SARS-CoV-2 and H1N1, and even variants. This is a major issue that can’t be readily addressed by current rapid tests."
The team continues working to further develop and test the technology with patient samples. Johns Hopkins Technology Ventures has applied for patents on the intellectual property associated with it and the team is pursuing license and commercialisation opportunities.