Shannon Cornett
Imaging Applications Manager, Bruker Daltonics
DOI: https://doi.org/10.1255/sew.2022.a12
© 2022 The Authors
Published under a Creative Commons BY-NC-ND licence
Clinical research has led the way in the use of matrix-assisted laser desorption/ionisation (MALDI) coupled with mass spectrometry imaging (MSI) in developing new applications to determine spatial data about biomolecules in tissues. MALDI imaging is a powerful, label-free analytical tool that can provide vital molecular information about protein modifications after gene expression. It also helps with visualising additional compounds like metabolites, glycans and lipids that play a part in disease pathology. Together with the emergence of new MALDI tools for targeted protein imaging, MALDI imaging offers the potential for scientists to fill in the gaps left by spatial transcriptomics and genomics in molecular investigations on tissue samples.1,2
This article describes MALDI imaging’s potential uses in pathology applications, and the benefits of the technique to map hundreds of biomolecules (proteins, lipids and glycans, for example) in a label-free, untargeted manner or for imaging target proteins using a modified immunohistochemistry protocol, often from a single tissue section. We also discuss the more recent influence of technological advances such as trapped ion mobility spectrometry (TIMS) and laser-induced post-ionisation (PI), as well as the impact on cancer research and disease management and how expanding the depth of available molecular information can translate into benefits for patient care.
Current use of biomolecule spatial data
The discovery of specific molecular markers of disease via molecular labels and probes has gradually expanded the traditional diagnostic histopathology of tissues. After the completion of the Human Genome Project in 2003, The Cancer Genome Atlas (TCGA) programme generated over 2.5 petabytes of genomic, epigenomic, transcriptomic and proteomic data from the molecular characterisation of over 20,000 primary cancer and matched normal samples, which spanned 33 cancer types. That data, which is available publicly, has already improved the ability to diagnose, treat and prevent cancer by connecting morphological information with genetic and molecular insights.
From these developments, the field of clinical molecular diagnostics emerged, aided by technological advancements such as sequencing technology, reverse transcription polymerase chain reaction (rt-PCR), immunohistochemistry (IHC) and fluorescence in situ hybridisation (FISH) methodologies. One perspective3 about these developments in the field summarised that: “Pathologists will become pilots for precision cancer therapy through their unique ability to combine morphological and molecular findings”. Indeed, the adoption of epidermal growth factor receptor (EGFR) mutation testing by pathology departments fits that narrative. By demonstrating the efficacy of kinase inhibitors erlotinib and gefitinib in the treatment of EGFR-mutated lung adenocarcinomas, the field’s paradigm shifted and began to emphasise the development of spatial omics techniques.
Current diagnostic tools
With the establishment of spatial information as a key component of analysing the microenvironment of disease in tissues, researchers turned to a suite of diagnostic tools such as IHC, spatial transcriptomics and imaging mass cytometry. While these tools serve multiple disease pathologies, which notably include neurology and cardiology, there is tremendous potential in the field of oncology where identifying the location and interactions of cellular components can dictate disease outcomes.
While these tools produce useful data about proteins in tissues, each one also brings challenges. The key limitations of all three tools include the inability to capture the variety of post-translational modifications (PTMs) in the cellular proteome, or to visualise the vast cellular lipidome and metabolome in context of tissue pathology, all of which would offer a broader molecular insight and stronger basis for classification compared to considering only the pre-translational proteome.
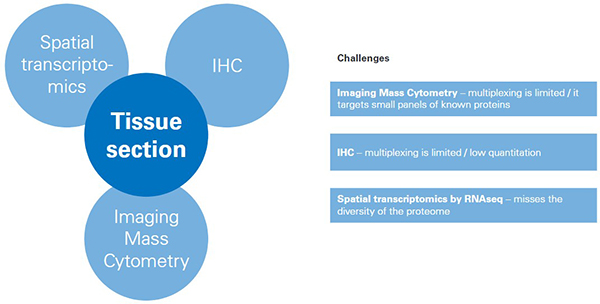
Figure 1. Challenges for tools for classifying/mapping cellular regions in tissue.
Impact of MALDI imaging
MALDI imaging is a powerful tool for mapping the distribution of molecules—ranging from small metabolites to large proteins—from a thin section of tissue without need for molecular tags or labels. A single MALDI imaging measurement can yield up to several thousands of distribution maps, or ion images, that reveal greater insight and understanding of molecular makeup and regional heterogeneity in the tissue. Due to its untargeted nature, it captures information about the spatial proteome and additional spatial omic signatures that are unique to the local cell neighbourhood without prior knowledge of the compounds. Many researchers agree that moving beyond protein biomarkers is important for next-stage clinical understanding, with one recent publication4 noting, for example, that: “Lipids play a significant role in the manifestation of cancer. However, research into lipid biomarkers of cancer is still in its infancy.”
For oncology applications, untargeted MALDI imaging enables the visualisation of tumour-associated biomolecules that are missed at the gene level. From a practical point of view, the MALDI imaging workflow is compatible with standard histological procedures, it maintains spatial resolution at around 10 µm, and the tissue section under test is preserved for further study. The result is a powerful complementary technique capable of discovering and spatially mapping important features at a deeper molecular level (Figure 2).
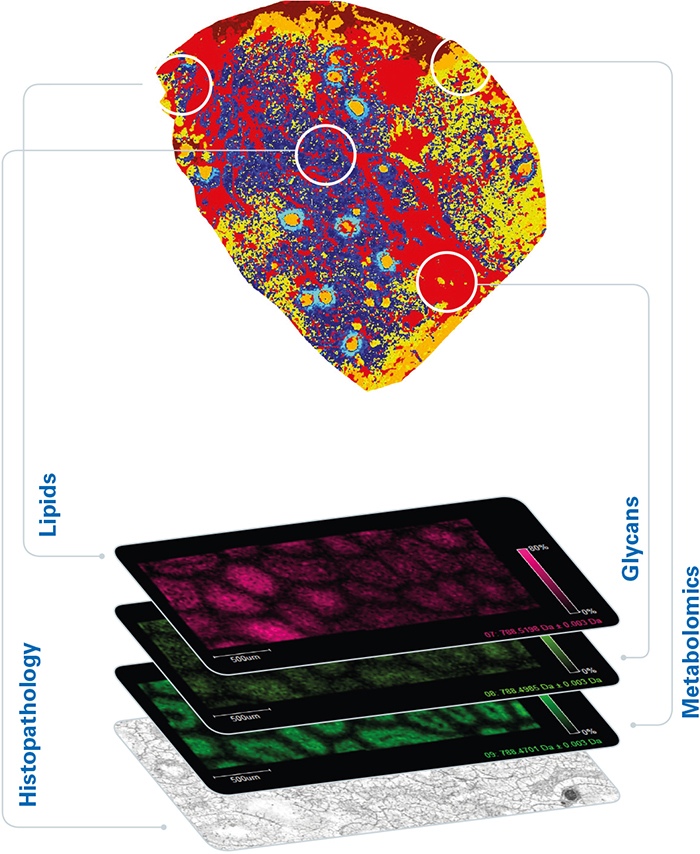
Figure 2. Additional disease insights available from MALDI imaging.
Routine oncology applications
MALDI imaging offers a great deal of potential for use in oncology. Detailed spatial proteomic, lipidomic and metabolomic insights can complement traditional genomic and transcriptomic methods. Often, knowing where molecular expression changes can be just as important as knowing if expression changes. This can be especially true if certain compounds are highly spatially concentrated or if molecules co-distribute in specific compartments because vital information is lost when examining only homogenised samples. The identification of new predictive or prognostic biomarkers, and the classification of heterogeneous tumour subpopulations, give important contextual clues to tissue-level communication networks that are integral to cancer growth and treatment success.
Many recent studies demonstrate the advantage of integrating MALDI imaging with traditional immuno techniques for tissue pathology applications. For example, Yagnik et al. reported the development of a new method based on novel photocleavable mass-tags (PC-MTs) for facile antibody labelling, which enables highly multiplexed IHC based on MALDI imaging (MALDI-HiPLEX IHC). The same technique, MALDI imaging, effectively projects untargeted imaging results into the familiar context of immuno-based cellular classification. The author’s conclusions were that the new combination shows promise for use in the fields of tissue pathology, tissue diagnostics, therapeutics and precision medicine.5
Work published in 2019 by Randall et al. demonstrated how MALDI imaging of lipids and metabolites in tissue samples accurately reflected a patient’s prostate cancer stage, as defined by traditional histologic evaluation using the “Gleason score”. This is the current standard of care; however, this time-consuming process is prone to intra/inter-observer variability, and provides no information about altered metabolic pathways or altered tissue architecture. They concluded that MALDI imaging could be used as a potential clinical tool to support more objective and faster diagnosis.6
To assess the suitability of MALDI imaging as a front-line technique, Bassu et al. recently reported on “real-time” molecular assessment of tumour margins. The goal was to discriminate surgical resection specimens from patients in a workflow that was rapid enough to directly impact surgical decisions.7 By adapting various stages of a conventional 30-minute protocol for MALDI imaging, they developed a reliable and reproducible 5-minute workflow that they concluded placed MALDI imaging firmly in the realm of routine clinical decision making (Figure 3). Furthermore, they suggest that by using an artificial intelligence (AI) step in the workflow, the MALDI imaging data could be analysed directly, without visual review, using previously established machine-trained models.
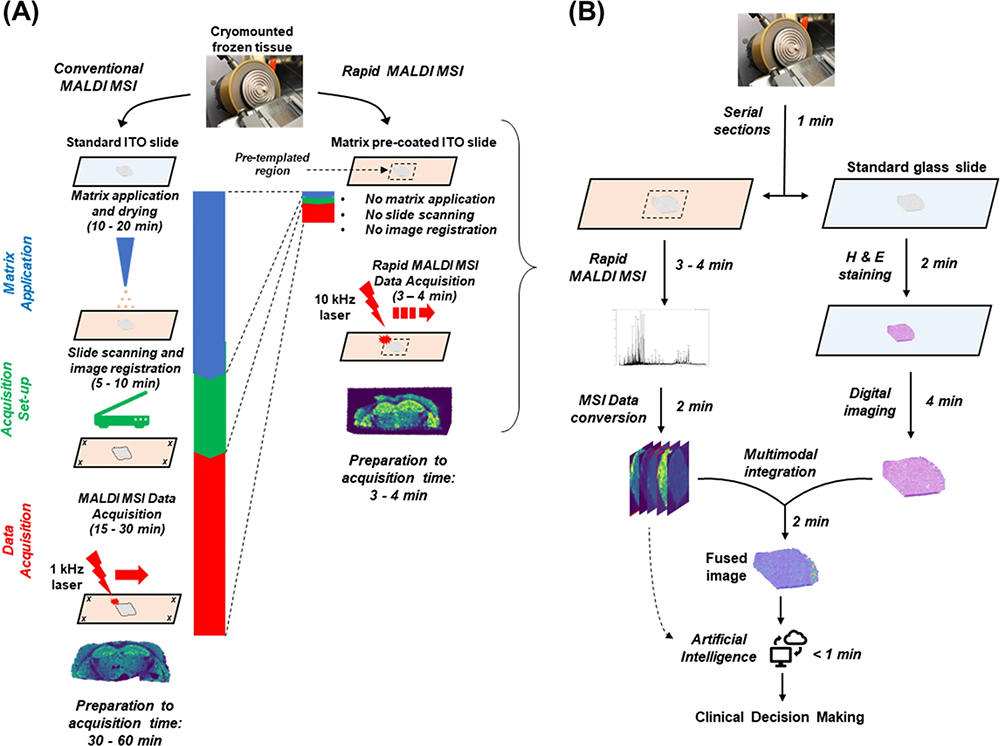
Figure 3. (A) Comparison of conventional vs rapid MALDI imaging. (B) Proposed workflow for MALDI imaging in the frozen section room. Figure reproduced from Reference 7 under a CC BY licence.
New technological advancements
Two recent breakthroughs in technology have significant implications for the future of MALDI imaging. First, ion mobility separation (IMS) has greatly broadened the range of biomolecules that can be analysed by pre-separation ahead of mass analysis. Of the many IMS technologies, Trapped IMS (TIMS) offers significant benefits for MALDI imaging as well as traditional omics. The timsTOF flex instrument (Bruker, Billerica, MA, USA) is one example, which adds a high spatial resolution MALDI imaging source to deliver a spatial dimension to omics analyses. The use of the technique was demonstrated in a joint project between Bruker and University of Maastricht, The Netherlands, that illustrated how MALDI-guided spatial omics uncovers proteomic diversity in lipid-segmented subpopulations of breast cancer that would otherwise have been missed using traditional bulk sampling.8
Second, novel laser-induced post-ionisation (PI) technology has delivered an increase in MALDI imaging sensitivity, by up to three orders of magnitude. Termed MALDI-2, this technology has now been applied to the timsTOF fleX and, together with TIMS, provides for the separation and mapping of even more analytes, such as lipids and glycans, in the complex tissue environment.9 Currently, this exciting development is being explored in research projects. However, it is easy to see how this technology could, in time, offer next-level performance to pathology applications too.
Conclusion
MALDI imaging provides significant advantages for molecular analysis of tissue specimens in greater detail. Crucially, MALDI imaging maintains the spatial relationships of analytes in tissues, allowing improved translational and clinical insights. That capability expands the accessibility of a broad spectrum of analytes in tissue including proteins, lipids, glycans and other analytes, in both targeted and untargeted manners.
The use of MALDI imaging offers new opportunities to create a top-down, disease-centric pathological view of tissues at the molecular level that can inform therapeutic strategies, support diagnosis and improve patient outcomes. Further developments to the technology will provide faster measurement speeds, increased sensitivity without compromising spatial resolution, and even deeper molecular content—important factors that will accelerate the adoption of MALDI imaging in the routine pathology environment.
References
- R.L. Caldwell and R.M. Caprioli, “Tissue profiling by mass spectrometry: a review of methodology and applications”, Mol. Cell. Proteomics 4(4), 394–401 (2005).
- A. Walch, S. Rauser, S.O. Deininger and H. Höfler, “MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology”, Histochem. Cell. Biol. 130, 421 (2008). https://doi.org/10.1007/s00418-008-0469-9
- L. Müllauer, “Milestones in pathology—from histology to molecular biology”, memo 10, 42–45 (2017). https://doi.org/10.1007/s12254-016-0307-z
- L.R. Stromberg, L.M. Lilley, H. Mukundan, “Advances in lipidomics for cancer biomarker discovery”, in Proteomic and Metabolomic Approaches to Biomarker Discovery (2nd Edn), Ed by H.J. Issaq and T.D. Veenstra. Academic Press, pp. 421–436 (2020). https://doi.org/10.1016/B978-0-12-818607-7.00025-6
- G. Yagnik, Z. Liu, K.J. Rothschild and M.J. Lim, “Highly multiplexed immunohistochemical MALDI-MS imaging of biomarkers in tissues”, J. Am. Soc. Mass Spectrom. 32(4), 977–988 (2021). https://doi.org/10.1021/jasms.0c00473
- E.C. Randall, G. Zadra, P. Chetta, B.G.C. Lopez, S. Syamala, S.S. Basu, J.N. Agar, M. Loda, C.M. Tempany, F.M. Fennessy and N.Y.R. Agar, “Molecular characterization of prostate cancer with associated Gleason score using mass spectrometry imaging”, Mol. Cancer Res. 17(5), 1155–1165 (2019). https://doi.org/10.1158/1541-7786.MCR-18-1057
- S.S. Basu, M.S. Regan, E.C. Randall, W.M. Abdelmoula, A.R. Clark, B. Gimenez-Cassina Lopez, D.S. Cornett, A. Haase, S. Santagata and N.Y.R. Agar, “Rapid MALDI mass spectrometry imaging for surgical pathology”, npj Precision Oncology 3, 17 (2019). https://doi.org/10.1038/s41698-019-0089-y
- J. Oetjen, R. Hebeler, C. Dewez et al., MALDI Guided SpatialOMx Uncovers Proteomic Profiles in Tumor Subpopulations of Breast Cancer. Application Note, Bruker Daltonics (2020). https://bit.ly/MSIAppNote
- J. Soltwisch, B. Heijs, A. Koch, S. Vens-Cappell, J. Höhndorf and K. Dreisewerd, “MALDI-2 on a trapped ion mobility quadrupole time-of-flight instrument for rapid mass spectrometry imaging and ion mobility separation of complex lipid profiles”, Anal. Chem. 92(13), 8697–8703 (2020). https://doi.org/10.1021/acs.analchem.0c01747

Shannon Cornett
Dale Shannon Cornett, PhD is a MALDI Imaging Market Manager at Bruker. He has over 30 years in the field of MALDI, having held numerous roles at Bruker, including Applications Scientist and Product Manager, and as Research Assistant Professor of Biochemistry at Vanderbilt University. Over the last 20 years he has worked with many leading researchers to develop new strategies and tools for using MALDI imaging mass spectrometry in pharmaceutical and clinical research.
[email protected]