Henry Rzepaa and Antony N. Daviesb
aDepartment of Chemistry, Molecular Sciences Research Hub, Imperial College London, White City Campus, Wood Lane, London W12 OBZ, UK
bSERC, Sustainable Environment Research Centre, Faculty of Computing, Engineering and Science, University of South Wales, UK
DOI: https://doi.org/10.1255/sew.2022.a10
© 2022 The Authors
Published under a Creative Commons BY licence
Ian Michael has challenged us on the topic of Open publishing: with all this focus on Findable data—where is it to be found? To get more data available in a form we can all use requires us all to re-think what we automatically do when we generate new spectra that might be of interest to fellow spectroscopists. Do we leave them to rot on an instrument computer, picking the “best of the best” to land in a peer reviewed publication some time years in the future? Or do we put procedures in place which ensure our work, and the work of our students, will be available to the scientific community for ever? Henry Rzepa has introduced a lovely educational spectroscopic and associated metadata workflow into Imperial College London. This not only ensures the students’ spectroscopic data and associated metadata is properly documented, but also made available in a FAIR way for the whole community. It also serves to introduce students to the whole concept of best practice in FAIR data handling and Persistent and Unique Identifiers, including the registration of the student to receive their own life-long unique publishing ORCiD ID.
How did Imperial College arrive at FAIR data publication by university students?
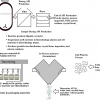
In 2012, the ORCiD organisation launched their researcher ID service, and shortly thereafter started to encourage research organisations to promote the concept to their researchers. At Imperial College, a debate was started to define the scope of who should be encouraged to register their ORCiD. One sensible, if predictable, plan was to start with the most senior research investigators, who often led large groups and to gradually work down to more junior members. A particular point of view that ended up not being adopted was to start at the other end of the researcher spectrum with undergraduate students, since they after all were the future of research! That latter option continued to resonate, however, and when the design for a new laboratory course for first year undergraduate students in the chemistry department at Imperial College in the first course year was started in 2018, the opportunity to enrol these students in ORCiD as part of that course was seized (see Figure 1, Step 1).
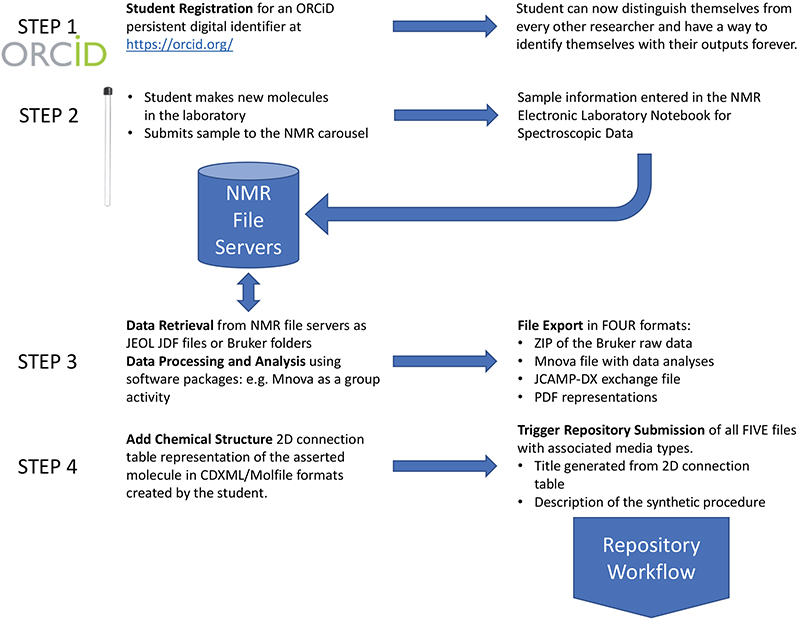
Figure 1. Student introduction to Persistent Identifiers through ORCiD ID registration and spectroscopic data generation and analysis of their work.
This was aided by the design of the course, which was to illustrate laboratory chemical synthesis by asking each student to generate a unique target molecule; more traditional courses involved a small number of previously known targets that all the students made. A careful choice of reaction allowed a large combinatorial design to be implemented in which two reagents (an alcohol and an acid) are combined to form an ester. Although each component is commercially available, the combination of the two would result in a molecule new to science. The next facet of the design was to ask the student to record spectroscopic information for this now unique and newly synthesised molecule, which would include the 1H NMR spectrum (the 13C was deemed too resource demanding for the spectrometer allocated to this course) along with an IR measurement. It was then a short step to encourage the student to formally publish this new data in a manner that the entire community of molecular scientists could potentially benefit from, by making this publication FAIR.1 This would mean that a simple search for the molecule by anyone based on its registered metadata (see below2) would be possible. Just as importantly, the FAIR attributes would allow a programmed and unsupervised machine to make the same searches, on any desired scale. The access to this data, its re-use by others and interoperability into new contexts was another benefit of this FAIR specification.
Rather than select a generic, non-subject specific repository such as Zenodo for this task, we already had access to a second generation FAIRsharing registered3 repository that had been custom-designed around rich “FAIR-Enabling” metadata4,5 and was already being used as the publication target for an ELN (electronic laboratory notebook) used by students undertaking computational laboratories.6,7
A simple but, at this stage, non-ELN based protocol was developed for the students to access, analyse and then publish their measured spectroscopic data in this repository.8 This experiment is now in its third year and the data acquired can be accessed via the repository collection.9
The FAIR publication workflow
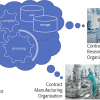
The additional workflows associated with this example of FAIR publishing of spectroscopic data is shown in Figure 2. Currently data publication comprises several distinct steps undertaken by the students, a process which could be automated in the future by introducing a spectroscopy-based ELN analogous to the system used for publishing data acquired in computational chemistry labs in which workflows can automatically generate required metadata.
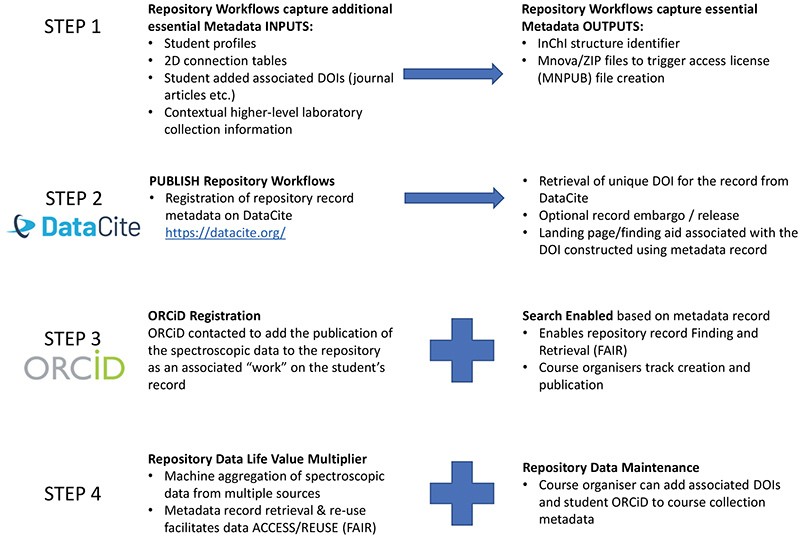
Figure 2. Spectroscopic Data Repository workflows showing interaction with DataCite for DOI generation, ORCiD for student data registration and the role in course delivery and monitoring.
A metadata model
Since this is about FAIR NMR data, it is useful to look at how the metadata record for an item imparts this aspect. This can take two forms, the media type10 of the data files uploaded to the repository and the properties of the data as registered in compliance with the DataCite schema. The properties are themselves usefully divided into a core set and a subject-specific set. DataCite core properties are fairly well established and include aspects such as the DOI identifier for the data record registration, the ORCiD(s) for the creators of the data, title, description, the affiliation identifier in the form of a ROR (Research Organisation Registry), the (re-use) rights identifier scheme, dates and “RelatedIdentifiers”, which can point to further metadata records of other “research objects” if they exist. Other RelatedIdentifiers can point to parent/children relationships involving other collections or datasets, other research objects such as associated journal articles and perhaps in the future instrument and even physical sample identifiers.
Currently the DataCite subject-specific metadata are currently less well formalised and standardised; an ongoing effort to achieve this for chemistry11 is under way under the auspices of IUPAC, the chemistry standards body. In 2016, some subject properties such as the InChI and InChI key chemical identifiers were included in the metadata collection by this repository.4,5 These are repository workflow generated from the molecular connection table submitted by the depositor (the CDXML file in the instance above). An example record can be downloaded using the link https://api.datacite.org/application/vnd.datacite.datacite+xml/10.14469/hpc/10176, where 10.14469/10176 is the DOI assigned to the data publication. Examples of how this can be exploited are shown below for you to try out.
Examples of FAIR spectroscopic data: how to find them
The metadata registration processes result in a so-called Aggregated Metadata Store (MDS). In this instance stored by DataCite, the selected registration authority. DataCite provide an API (Application Programming Interface) to search the store and they plan to introduce a more human-friendly form. The API calls are relatively human “readable” and a number of them are documented being pertinent for the student data publication project.2 They typically operate by specifying the property as defined in the metadata record and its required value (a key-pair) and using Boolean logic can be combined with the media type values to focus the search.
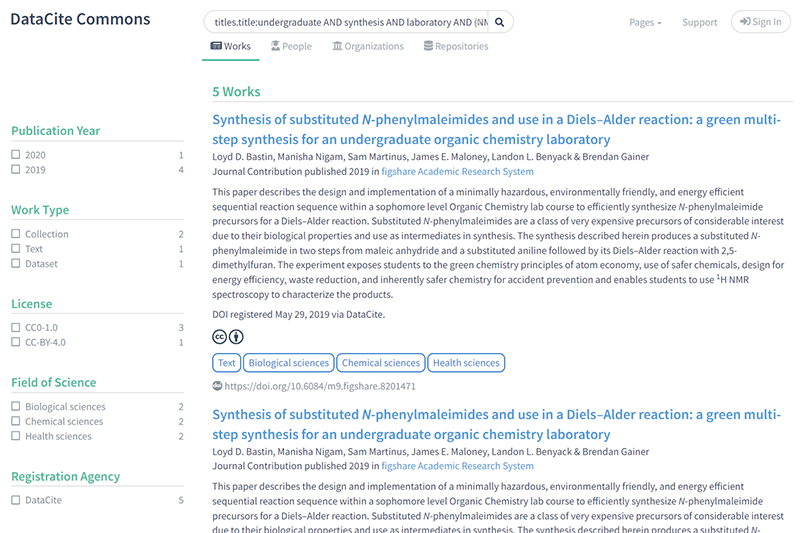
1) This example is based on using keywords present in the title or description of an entry: https://commons.datacite.org/?query=titles.title:undergraduate+AND+synthesis+AND+laboratory+AND+(NMR+OR+SPECTRUM) which as we go to press reveals five hits. Three are associated with the course being discussed here and two are from another course. The first three are identified from the metadata as collections of items, the collections pointing to “children” as actual datasets.
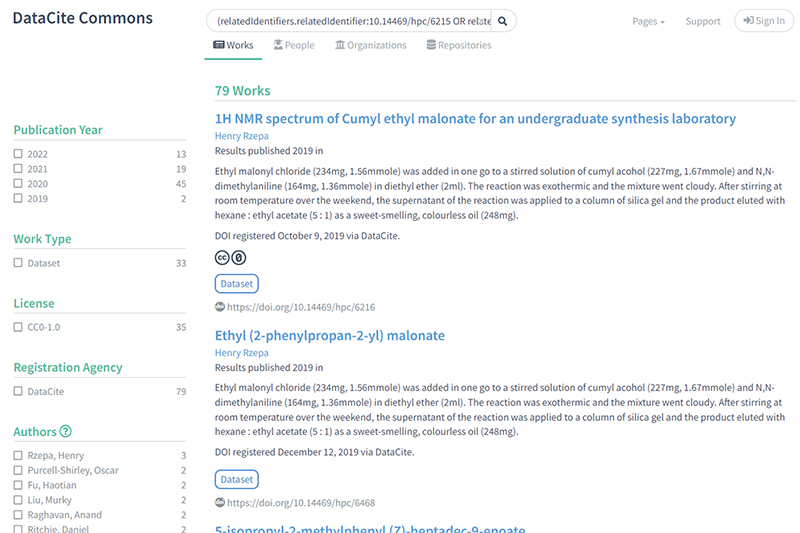
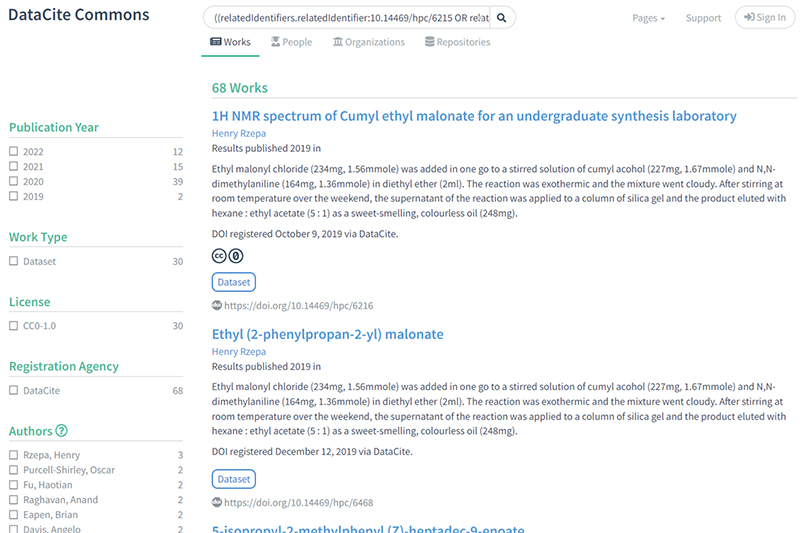
2) These datasets can be found by searching specifically on the three collections identified in the previous search to find the associated children: https://commons.datacite.org/?query=(relatedIdentifiers.relatedIdentifier:10.14469/hpc/6215+OR+related Identifiers.relatedIdentifier:10.14469/ hpc/7350+OR+related Identifiers.relatedIdentifier:10.14469/hpc/8679)+AND+relatedIdentifiers.relationType:IsPartOf gives 79 works.
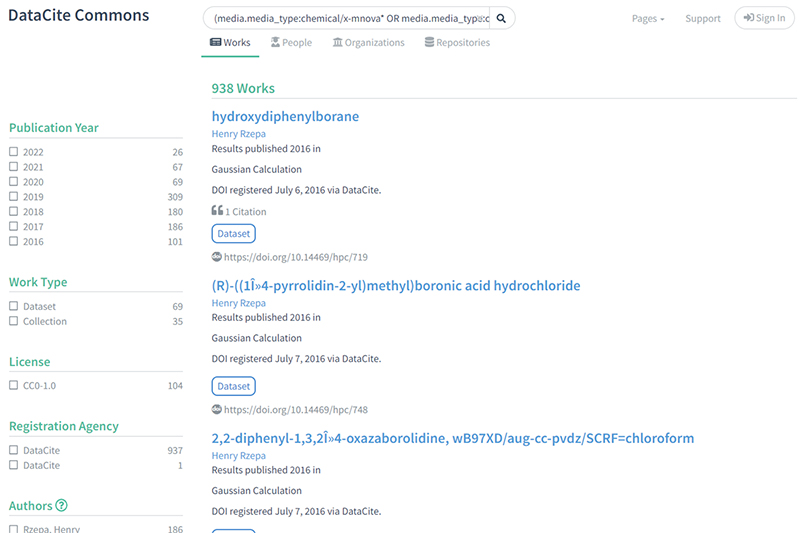
3) This search adopts a different strategy, which is to search for all datasets with media types associated with NMR data, which in fact returns the globally aggregated set having these metadata properties: https://commons.datacite.org/?query=(media.media_type:chemical/x-mnova*+OR+media.media_type:chemical/x-jeol-jdf+OR+media.media_type:chemical/x-jcamp-dx+OR+(chemical/x-mnpub*+AND+media.media_type:application/zip))
4) Searches 2 and 3 could be combined into a single search using a Boolean operator (red) to verify that the children previously identified in search 2 actually contain NMR datasets: https://commons.datacite.org/?query=((relatedIdentifiers.relatedIdentifier:10.14469/hpc/6215+OR+relatedIdentifiers.relatedIdentifier:10.14469/hpc/7350+OR+relatedIdentifiers.relatedIdentifier:10.14469/hpc/8679)+AND+relatedIdentifiers.relationType:IsPartOf)+AND+(media.media_type:chemical/x-mnova*+OR+media.media_type:chemical/x-jeol-jdf+OR+media.media_type:chemical/x-jcamp-dx+OR+(chemical/x-mnpub*+AND+media.media_type:application/zip))
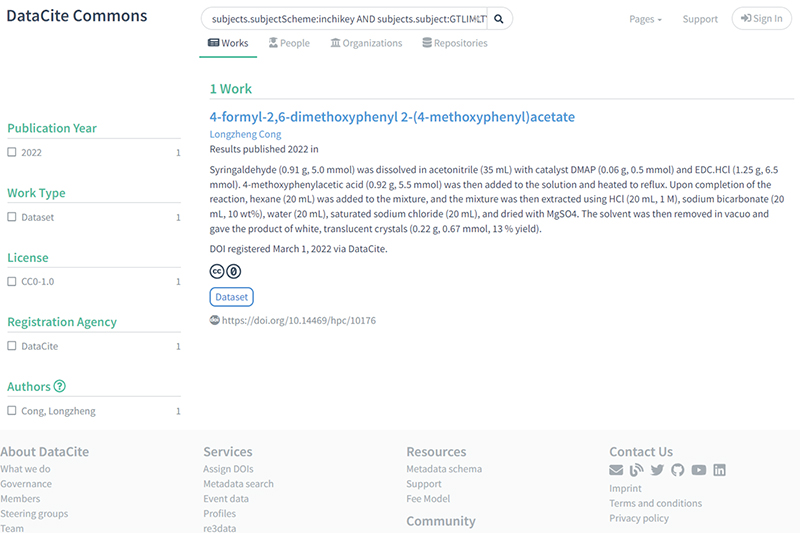
5) The next search illustrates hunting for a very specific molecule using its InChI string and of course this search could be combined with other searches if desired using Booleans: https://commons.datacite.org/?query=subjects.subjectScheme:inchikey+AND+subjects.subject:GTLIMLTYVRBPEP-UHFFFAOYSA-N
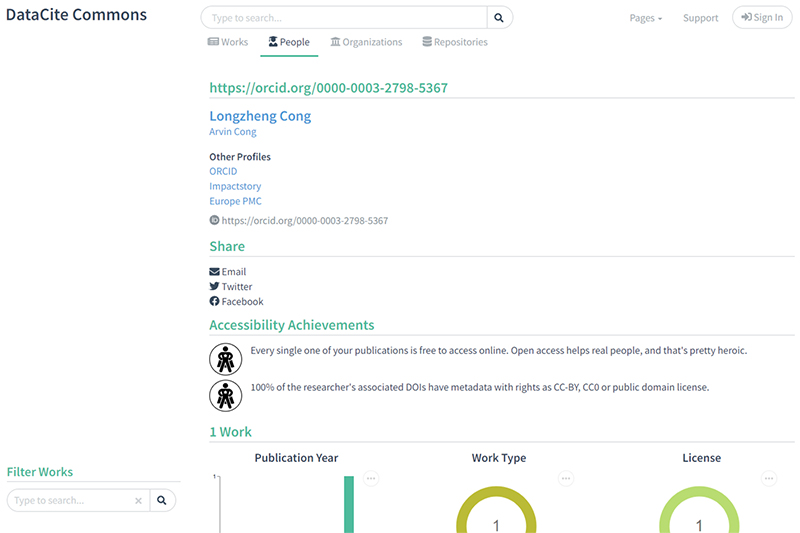
6) Clicking on the author’s name in Search 5 (Longzheng Cong) leads to a summary page of the author’s submissions to the DataCite repository. https://commons.datacite.org/orcid.org/0000-0003-2798-5367
The above illustrate a selection of potential searches only. These could be combined as desired with other metadata properties such as author (specified by ORCiD), institution (specified by ROR, journal or specific journal article (specified by DOI prefix) and associations with other types of data such as computation, or other types of spectroscopy such as IR spectra or crystal structures etc. The possibilities are vast.
Examples of FAIR spectroscopic data: how to access and re-use them
Following up the result of, say search 5 above, reveals a collection of NMR-related files, in different expressions of the data or different representations of the spectrum. A human can select any of these files for download but is then dependent on specialised software to open the raw data files (in this case the Jeol JDF file or a processed Mnova file). This would normally require possession of a software license allowing access to the program, which might limit access.
To address this specific issue, a further file has been generated by the Imperial repository which provides a free-to-use license to grant access to the data; the MNPUB file. Downloading the MNPUB files(s) specifically grants the user access to the full functionality of the Mnova program to process that dataset. This reinforces one aspect of the A in FAIR; access is not simply about being able to download a dataset but being able to use it in appropriate software to expose the full information the data carries.
FAIR is, however, not just about granting access to people; what about machines? Hidden behind the scenes in the metadata is the following declaration just for them:
<relatedIdentifier relatedIdentifierType=”URL” relationType=”HasMetadata”>https://data.hpc.imperial.ac.uk/resolve/?ore=10176</relatedIdentifier>
This specifies how a machine might acquire what might be called a file manifest of further metadata. This manifest contains information on the media types for the files and how to access them, which the machine can exploit.
The future
Clearly the example which Henry has detailed above delivers far more than just a spectroscopic data repository workflow. The students’ learnings from such an exercise go way beyond the creation of spectroscopic data. The work shows the importance of the persistent identifiers not only for the data and metadata but also, through the ORCiD ID, of their identity as full members of the scientific community.
This work also hammers home the critical and essential importance of carefully created and curated metadata, as much of the functionality illustrated above is achieved with the help of metadata. Importantly, it also depends on community agreement on how that metadata is specified. As noted earlier, an IUPAC working party is expected to produce recommendations on these aspects soon.11 The examples above were produced as proofs-of-concept, but in due course will be harmonised with the emerging recommendations. The next challenge is to encourage all data repositories which contain, for example, NMR spectroscopic data to also adopt these recommendations. When this happens, much richer chemical metadata allowing much more specific searches than the ones above will become enabled.
Acknowledgements
We thank Dr Ed Smith for the design and implementation of the synthetic experimental aspects.
References
- M.D. Wilkinson, M. Dumontier, […] B. Mons, Sci. Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18
- H.S. Rzepa and S. Kuhn, Mag. Res. Chem. 60, 93–103 (2021). https://doi.org/10.1002/mrc.5186
- FAIRsharing.org: Imperial College Research Data Repository. https://doi.org/10.25504/FAIRsharing.LEtKjT
- J. Downing, P. Murray-Rust, A.P. Tonge, P. Morgan, H.S. Rzepa, F. Cotterill, N. Day and M.J. Harvey, J. Chem. Inf. Mod. 48, 1571–1581 (2008). https://doi.org/10.1021/ci7004737
- M.J. Harvey, A. McLean and H.S. Rzepa, J. Cheminform. 9, 4 (2017). https://doi.org/10.1186/s13321-017-0190-6
- M.J. Harvey, N.J. Mason and H.S. Rzepa, J. Chem. Inf. Model. 54, 2627–2635 (2014). https://doi.org/10.1021/ci500302p
- C. Cave-Ayland, M.J. Bearpark, C. Romain and H.S. Rzepa, “CHAMP is a HPC access and metadata portal”, J. Open Source Soft. 7(70), 3824 (2021). https://doi.org/10.21105/joss.03824
- H.S. Rzepa, Publishing NMR Research Data. https://doi.org/10.14469/hpc/6472
- Undergraduate Synthesis Laboratories at Imperial College. Imperial College Research Data Repository. https://doi.org/10.14469/hpc/7349
- H.S. Rzepa, P. Murray-Rust and B.J. Whitaker, J. Chem. Inf. Comp. Sci. 38, 976–982 (1998). https://doi.org/10.1021/ci9803233
- R.M. Hanson, D. Jeannerat, M. Archibald, I. Bruno, S.J. Chalk, A.N. Davies, R.J. Lancashire, J. Lang and H.S. Rzepa, Pure Appl. Chem., in press (2022). https://doi.org/10.1515/pac-2021-2009

Henry Rzepa
Henry Rzepa was trained as an experimental physical organic chemist and then spent three years learning the emerging area of computational chemistry with Michael Dewar in Austin, Texas. Upon joining the staff at Imperial College in 1977, his researches became focussed on computational mechanistic chemistry, NMR and chiroptical spectroscopies and Internet-based Chemical informatics, for which he was awarded the 2012 ACS Skolnik award. 0000-0002-8635-8390
0000-0002-8635-8390
[email protected]

Tony Davies
Tony Davies is a long-standing Spectroscopy Europe column editor and recognised thought leader on standardisation and regulatory compliance with a foot in both industrial and academic camps. He spent most of his working life in Germany and the Netherlands, most recently as Lead Scientist, Strategic Research Group – Measurement and Analytical Science at AkzoNobel/Nouryon Chemicals BV in the Netherlands. A strong advocate of the correct use of Open Innovation. 0000-0002-3119-4202
0000-0002-3119-4202
[email protected]













Search 6 above
To correct a minor statement, Clicking on the author’s name in Search 5 (Longzheng Cong) leads to a summary page of the author’s submissions to the DataCite repository, it is the author's submissions to ANY repository where their ORCID is present in the metadata record that is recorded by DataCite.
Caption to search 6.
The caption with the text Clicking on the author’s name in Search 5 (Longzheng Cong) leads to a summary page of the author’s submissions to the DataCite repository. should be changed to Clicking on the author’s name in Search 5 (Longzheng Cong) leads to a summary page of all the author’s submissions to any repository for which metadata has been registered with DataCite.