Robert M. Hanson,a Damien Jeannerat,b Mark Archibald,c Ian Bruno,d Stuart Chalk,e Antony N. Davies,f Robert J. Lancashire,g Jeffrey Langh and Henry S. Rzepai
aDepartment of Chemistry, St Olaf College, Northfield, Minnesota, USA
bNMRprocess.ch, Geneva, Switzerland
cRoyal Society of Chemistry, Cambridge, UK
dCambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK
eDepartment of Chemistry, University of North Florida, Jacksonville, FL, USA
fSERC, Sustainable Environment Research Centre, Faculty of Computing, Engineering and Science, University of South Wales, UK
gProf. Emeritus, Department of Chemistry, The University of the West Indies, Kingston 7, Mona Campus, Jamaica
hAmerican Chemical Society Publications Division, Rockville, Maryland, USA
iDepartment of Chemistry, Imperial College London, Molecular Sciences Research Hub, White City Campus, 82 Wood Lane, London W12 0BZ, UK
DOI: https://doi.org/10.1255/sew.2021.a9
© 2021 The Authors
Published under a Creative Commons BY licence
As 2021 arrives with all the promise of beating COVID-19, mixed with the realities of vaccine delays and the threatened breakdown of international consensus on tackling global pandemics, it is a strange feeling to be writing an article about the ability of all to find, access, understand and re-use spectroscopic data. However, effectively and freely sharing information needs to be at the heart of any global effort to beat a pandemic. Although nationalistic tendencies always raise their ugly heads at times of tension, it is only through strong global collaboration and free access to data that we can hope to minimise the damage to our nearest and dearest!
The International Science Council report on Opening the Record of Science
February 2021 has just seen the publication of the ISC Report on Opening the Record of Science: making scholarly publishing work for science in a digital era Figure 1.1

Figure 1. The ISC Report on Opening the Record of Science.
A PDF version of the report is appropriately free to download for those who want to read it in full at https://council.science/wp-content/uploads/2020/06/2020-02-19-Opening-the-record-of-science.pdf. One of the things I like about this report is that it starts with the basics, briefly explaining “Why Science Matters” including the clear statement around the communication of new experiments and new observations publicly communicated through the published record of science. The importance of the publication process is nicely summed up with “Publication processes that achieve these ends and are adapted to the needs and priorities of the disciplines of science and interdisciplinary collaboration are essential to the function of science as a global public good”.
The ISC Report lays down seven Principles for Scientific Publishing around how modern scientific publishing needs to serve us, and which need be durable in the long term:
- There should be universal open access to the record of science, both for authors and readers.
- Scientific publications should carry open licenses that allow reuse and text and data mining.
- Rigorous and ongoing peer review is essential to the integrity of the record of science.
- The data/observations underlying a published truth claim should be concurrently published.
- The record of science should be maintained to ensure open access by future generations.
- Publication traditions of different disciplines should be respected.
- Systems should adapt to new opportunities rather than embedding inflexible infrastructures.
Who is the ISC?
The International Science Council (ISC) is a non-governmental organisation with a global membership consisting of 40 international scientific Unions and Associations and over 140 national and regional scientific organisations including Academies and Research Councils.
For spectroscopists, the Principles around Open Access, Reuse of the Text and the Data and the need to concurrently publish both reinforce the position this column has taken from its inception. We do not have space to go through the whole Report, but it is well worth highlighting their approach to the importance of scientific data, including a very powerful statement that “Publishing the data is as important, and sometimes more important, than publishing the written text”. The ISC2 specifically refer to the FAIR data principles which we have covered in a few articles, including reporting on the IUPAC CODATA Workshop on “Supporting FAIR Exchange of Chemical Data through Standards Development” held in Amsterdam in July 2018,3 which led to the founding of an IUPAC project in 2019.
IUPAC Project “Development of a standard for FAIR data management of spectroscopic data”
The objective of this project is to apply FAIR data principles to spectroscopic data in the field of chemistry building on IUPAC’s extensive expertise in this area. The project will develop standards for the production and dissemination of digital data objects that contain enough spectral data and metadata that they can be:
- findable through semantic searches on the web,
- available through standard interfaces,
- interoperable and transferable between systems and
- readable and reusable over time, for both humans and machines.
The IUPAC FAIRSpec Project
Following the Amsterdam workshop, a proposal was submitted to IUPAC for a project to follow up on many of the actions agreed at the workshop. The project, under the title of Development of a Standard for FAIR Data Management of Spectroscopic Data, was launched under the auspices of the IUPAC Committee on Publications and Cheminformatics Data Standards right at the start of the COVID epidemic on 18 March 2020 (https://iupac.org/project/2019-031-1-024). The project objectives are shown in the textbox. There has been a lot of work done this year around exactly what role IUPAC, as the standardisation body for chemistry, can play in the FAIR initiative. Clearly IUPAC’s ownership and responsibility for many cheminformatics data standards, especially the JCAMP-DX series of spectroscopic data standards, places IUPAC in a special position to respond when the environment for which these standards were originally crafted changes radically. FAIR is just such a ground-breaking change, where the correct storage and accessibility with data processing opportunities means that the “minimum essential metadata” approach of the original standards needs to be brought up to date by standardising the majority of the relevant metadata with a spectroscopic data set. Indeed, this column challenged the spectroscopic community in a relatively recent article called “Are you taking your Metadata seriously?”.4
The project team have produced a useful figure as they continue to assess the current state of available data collections. It is reproduced in Figure 2 for the example of NMR data.
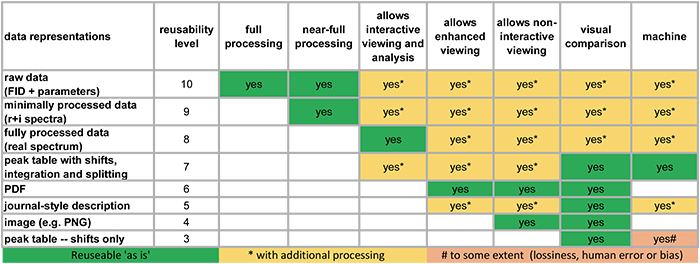
Figure 2. How reusable are our NMR data in scientific publications.
One advantage of IUPAC projects being able to draw on some of the top innovators in the field is that we have people who are actively FAIRifying their own working environments. The following sections will show a few examples of how this work is progressing.
Example of accompanying spectroscopic data from ACS Publications: submission of real data (Jeff Lang)
In February of 2020, ACS Publications began a programme to encourage authors to submit their original data for NMR, including free induction decay (FID) files, acquisition data and processing parameters in a zip file as Supporting Information with their manuscript at submission time. Two journals, The Journal of Organic Chemistry and Organic Letters joined this programme by publishing a joint editorial.5 The goal was to utilise existing scholarly infrastructure to encourage data publication and gauge support for data publication from the chemistry community.
In the first year of the programme, these two journals published nearly 200 manuscripts with NMR primary data, demonstrating early support. These data are available during manuscript review and receive a DOI upon publication. ACS provided a tool for authors to package the primary data with metadata like structure identifiers, ORCID and funding identifiers. These would better align the resulting package with the FAIR data principles,6 but authors have preferred to package the data themselves, often forgoing such metadata. Future efforts will focus on the incentives and workflow needed to solicit this metadata in a scalable way.
Example of accompanying spectroscopic data from the Royal Society of Chemistry: ChemSpider (Mark Archibald)
ChemSpider contains approximately 400,000 community-submitted NMR, IR, UV-vis and mass spectra (most come from existing collections or projects). Although this is a significant set of spectra, it represents a tiny percentage of >100 million total ChemSpider records. ChemSpider records can be found by searching on structure, text, experimental or calculated properties, or combinations thereof. However, it is not currently possible to limit a search to only records containing spectra—the user must click through to the record to discover if a spectrum is present. Spectra within ChemSpider records are freely available to all users (Figure 3 is an example ChemSpider entry for acetaldehyde). At present, the ChemSpider APIs do not enable retrieval of spectra, limiting opportunities for machine processing. The majority (~300,000) are in JCAMP format, so they rate highly for reusability. An interactive viewer (JSpecView) allows visualisation of JCAMP spectra on the record page.
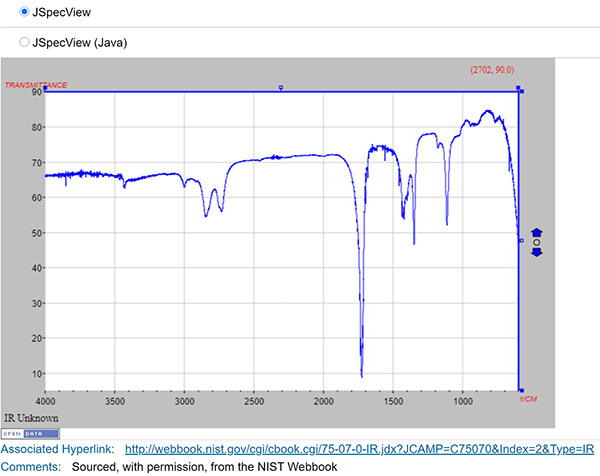
Figure 3. ChemSpider data example.
Using standardised spectroscopic metadata to facilitate cross-continent data enhancement workflows (Robert Hanson)
In discussions within the project, it has become clear that the chemical structure representation may well be one of the most important “metadata” objects associated with spectra. In an earlier column,7 a scheme was introduced whereby an NMR spectrum could be predicted (initially 1H, but now 13C as well) and the input could be a name of the compound or a 2D structure drawn with JSME. This relied on the generation of sufficient information that could be forwarded to nmrdb.org at the École Polytechnique Fédérale de Lausanne (EPFL) for processing (Figure 4).8–10
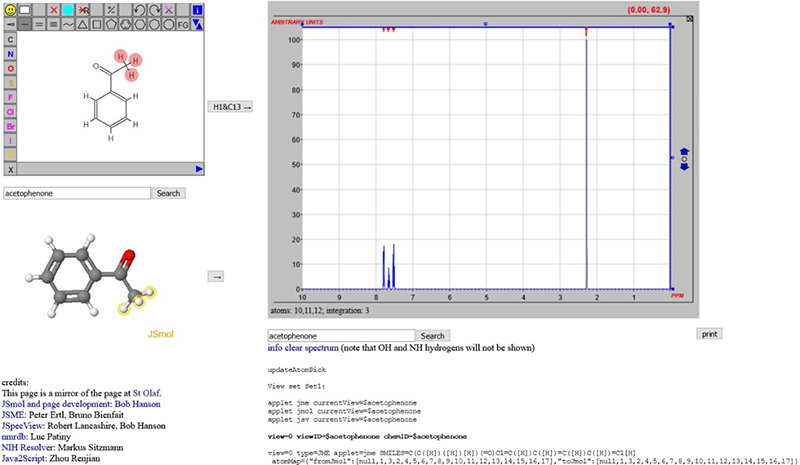
Figure 4. From structures to spectra using trans-continental data processing.
Chemical structure metadata, such as connection tables between the atoms or simply the chemical name input, allows the processing to begin, as Table 1 shows.
Table 1. Path of information exchange starting from a structure and from a name.
From drawing a structure: | |
JSME > SMILES (no H atoms) | local |
SMILES > NCI > 2D SDF file (with H atoms) | USA |
2D SDF file > JSME for display |
|
SMILES > NCI > 3D SDF file | USA |
From typing a name: | |
name > NCI > 2D SDF file (with H atoms) | USA |
2D SDF file > JSME for display | local |
name > NCI > 3D SDF file | USA |
Then from either method: | |
3D SDF file to JSmol and sent to EPFL | local |
Generates modified 3D mol file, sends to Lisbon | Switzerland |
Generates chemical shift+coupling+atom correlation matrix, sent to EPFL | Portugal |
EPFL applies a second-order coupling algorithm and line broadening then sends spectral data, assignment and (modified) 3D mol file | Switzerland |
2D and two 3D model atom numbering is reconciled using fully elaborated SMILES matching.* Annotated JCAMP-DX file and 3D mol displayed. Interactive atom/peak selection is enabled. | local |
*see https://chemapps.stolaf.edu/jmol/jsmol/correlate.htm
NMRDB references
The following services are available, compatible with HTML5, where a SMILES string is embedded in the call.
1H NMR prediction: https://www.nmrdb.org/service.php?name=nmr-1h-prediction&smiles=c1ccccc1CC
13C NMR prediction: https://www.nmrdb.org/service.php?name=nmr-13c-prediction&smiles=c1ccccc1CC
COSY prediction: https://www.nmrdb.org/service.php?name=cosy-prediction&smiles=c1ccccc1CC
HSQC/HMBC prediction: https://www.nmrdb.org/service.php?name=hmbc-prediction&smiles=c1ccccc1CC
All predictions: https://www.nmrdb.org/service.php?name=all-predictions&smiles=c1ccccc1CC
1H NMR prediction was possible thanks to the tool of the FCT-Universidade NOVA de Lisboa developed by Yuri Binev and Joao Aires-de-Sousa.11
Incorporating FAIR principles into undergraduate teaching (Henry Rzepa)
As a final example, Henry Rzepa has been developing a novel approach to capturing and disseminating NMR spectroscopic data, which incorporates the basic FAIR principles into an undergraduate student experiment illustrating the synthesis of an organic ester from carboxylic acid and phenolic components.12
Each student in a year class is assigned a different combination of reactants, chosen so that the resulting synthesis produces a new-to-science molecule. Following workup, the student submits the sample for NMR analysis and the resulting instrument dataset is acquired/processed to produce a spectrum from the raw FID. The students then participate in group analysis of their individual spectra.
Finally, the student moves to publishing the primary instrumental data in a FAIRsharing data repository13–14 in the form of both a ZIP archive and processed versions also containing an annotated spectrum (MestreNova archive + JCAMP-DX). The student adds further core metadata to the record and initiates workflows which include generating a chemical identifier (InChI string and key) and a free-to-use MestreNova dataset Access license.15
Publication produces a metadata record registered against a persistent identifier (DOI),16 the latter eventually appearing in the student’s ORCID researcher profile. The metadata has sufficient information to allow a variety of complex Finding searches to be undertaken12 and includes Access information allowing potentially unsupervised machine Interoperation and Reuse of the data for e.g. further AI-based spectral analysis.12
Conclusions
So, we already do have some good examples of interoperability between diverse systems, but, as you can see from the examples above, they are pretty much all reliant on the developers’ knowledge of the specifics of the raw spectroscopic data sets they receive, as opposed to being able to call on standardised definitions for the metadata in spectroscopy outside of the limited use in JCAMP-DX standards.
Everyone please, stay safe!
References
- International Science Council, Opening the Record of Science: Making Scholarly Publishing Work for Science in the Digital Era. International Science Council, Paris, France (2021). https://doi.org/10.24948/2021.01
- https://council.science/about-us/
- L. McEwen, D. Martinsen, R. Lancashire, P. Lampen and A.N. Davies, “Are your spectroscopic data FAIR?”, Spectrosc. Europe 30(4), 21–24 (2018). https://doi.org/10.1255/sew.2018.a2
- A.N. Davies, P. Lampen and R. Lancashire, “Are you taking your Metadata seriously”, Spectrosc. Europe 31(2), 17–23 (2019). https://doi.org/10.1255/sew.2019.a1
- A.M. Hunter, E.M. Carreira and S.J. Miller, “Encouraging submission of FAIR data at The Journal of Organic Chemistry and Organic Letters”, Org. Lett. 22(4), 1231–1232 (2020). https://doi.org/10.1021/acs.orglett.0c00383; A.M. Hunter, E.M. Carreira and S.J. Miller, “Encouraging submission of FAIR data at The Journal of Organic Chemistry and Organic Letters”, J. Org. Chem. 85(4), 1773–1774 (2020). https://doi.org/10.1021/acs.joc.0c00248
- M.D. Wilkinson, M. Dumontier, I. Aalbersberg et al., “The FAIR Guiding Principles for scientific data management and stewardship”, Sci. Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18
- A.N. Davies, M. Cashyap, R. Lancashire and R.M. Hanson, “A head in the clouds?—Part two: exploring distributed, multi-server 1H NMR prediction”, Spectrosc. Europe 26(5), 15–17 (2014). https://doi.org/10.1255/sew.2014.a1
- D. Banfi and L. Patiny, “www.nmrdb.org: Resurrecting and processing NMR spectra on-line, Chimia 62(4), 280–281 (2008). https://doi.org/10.2533/chimia.2008.280
- A.M. Castillo, L. Patiny and J. Wist, “Fast and accurate algorithm for the simulation of NMR spectra of large spin systems”, J. Magn. Reson. 209(2), 123–130 (2011). https://doi.org/10.1016/j.jmr.2010.12.008
- J. Aires-de-Sousa, M. Hemmer and J. Gasteiger, “Prediction of 1H NMR chemical shifts using neural networks”, Anal. Chem. 74(1), 80–90 (2002). https://doi.org/10.1021/ac010737m
- Y. Binev, M.M. Marques, J. Aires-de-Sousa, “Prediction of 1H NMR coupling constants with associative neural networks trained for chemical shifts”, J. Chem. Inform. Model. 47(6), 2089–2097 (2007). https://doi.org/10.1021/ci700172n
- H.S. Rzepa and S. Kuhn, submitted for publication (2021).
- FAIRsharing.org, Imperial College Research Data Repository. https://doi.org/10.25504/FAIRsharing.LEtKjT
- A.N. Davies, D. Martinsen, H.S. Rzepa, C. Romain, A. Barba, F. Seoane, S. Dominguez and C. Cobas, “Simplifying spectroscopic supplementary data collection”, Spectrosc. Europe 29(4), 6–8 (2017). https://doi.org/10.1255/sew.2017.a1
- A. Barba, S. Dominguez, C. Cobas, D.P. Martinsen, C. Romain, H.S. Rzepa and F. Seoane, “Workflows allowing creation of journal article supporting information and Findable, Accessible, Interoperable, and Reusable (FAIR)-enabled publication of spectroscopic data”, ACS Omega 4(2), 3280–3286 (2019). https://doi.org/10.1021/acsomega.8b03005
- H.S. Rzepa et al., “1st Year undergraduate synthesis laboratory (2019–2020)”, Imperial College London data repository (2020). https://doi.org/10.14469/hpc/6215

Bob Hanson
Bob Hanson is a professor in the Chemistry Department at St Olaf College. He is one of the principal developers of Jmol, JSmol,and JSpecView. 0000-0001-5411-2356
0000-0001-5411-2356
[email protected]

Damien Jeannerat
Damien Jeannerat is an independent chemical data specialist. He first worked on the development of nuclear magnetic resonance (NMR) applied to organic compounds and natural products of pharmacological interest. Followed, twenty years of academic research at the University of Geneva where funded and coordinated the NMReDATA Initiative which introduced a format and method to share the NMR information with the community. After leaving academic research, he works for CHEMeDATA.org, co-chairs at IUPAC working group developing recommendations for spectroscopic data and looks for new fruitful collaborations aiming at actively increase the FAIRness of chemistry data. 0000-0001-7018-4288
0000-0001-7018-4288
[email protected]

Mark Archibald
In nine years at the Royal Society of Chemistry, Mark Archibald has worked on the journal Organic & Biomolecular Chemistry, The Merck Index Online, the National Chemical Database Service and most recently ChemSpider. He applies his background in synthetic organic chemistry to the curation and bulk processing of chemical data in ChemSpider and is interested in cheminformatics and data management more broadly. 0000-0001-8687-7134
0000-0001-8687-7134
[email protected]

Ian Bruno
Ian Bruno is Director of Data Initiatives at the Cambridge Crystallographic Data Centre which has been managing and curating scientific data for over 50 years. Ian himself has over 25 years’ experience in the world of chemistry and informatics. He is an active contributor to data-related activities within IUPAC and is Secretary of the InChI Trust.
[email protected]

Stuart J. Chalk
Dr Stuart J. Chalk is a Professor in the Department of Chemistry at the University of North Florida. Although trained as an analytical chemist, Dr Chalk’s research now focuses on the areas of Chemical Informatics and Data Science. In particular, Dr Chalk has projects focused on machine accessibility of solubility online enhancement to the IUPAC Gold Book, automated extraction and annotation of chemical property data from PDF files and scientific data models. Dr Chalk’s newest NSF funded grant focuses on semantic integration of heterogeneous datasets from toxicology, medicine, materials, biodiversity, and chemistry. 0000-0002-0703-7776
0000-0002-0703-7776
[email protected]

Tony Davies
Tony Davies is a long-standing Spectroscopy Europe column editor and recognised thought leader on standardisation and regulatory compliance with a foot in both industrial and academic camps. He spent most of his working life in Germany and the Netherlands, most recently as Lead Scientist, Strategic Research Group – Measurement and Analytical Science at AkzoNobel/Nouryon Chemicals BV in the Netherlands. A strong advocate of the correct use of Open Innovation. 0000-0002-3119-4202
0000-0002-3119-4202
[email protected]

Robert Lancashire
Prior to his retirement in 2015, Robert Lancashire was the Professor of Computational Chemistry at the Department of Chemistry, The UWI, Mona. He joined the staff there in 1979 and held the positions of Sub-Dean for Technology from 1998 to 2004 and Deputy Dean for 2007–2008. He was conferred Professor Emeritus in 2016. He served as secretary of the International Union of Pure and Applied Chemistry (IUPAC) Committee on Printed and Electronic Publications (CPEP). 0000-0002-6780-3903
0000-0002-6780-3903
[email protected]
Jeff Lang
Jeff Lang is Assistant Director for Platform Development at The American Chemical Society Publications Division, where he supports innovation and product development that explores the untapped potential of technology to enhance scholarship. Prior to this role, Jeff worked at ProQuest in many departments, including Product Owner for RefWorks and Community Manager of GradShare.com a graduate student support community. Jeff has degrees in Computer Science and Information Management and has spent the last 20 years at the nexus of technology and academic publishing services. 0000-0003-4895-0278
0000-0003-4895-0278
[email protected]

Henry Rzepa
Henry Rzepa was trained as an experimental physical organic chemist and then spent three years learning the emerging area of computational chemistry with Michael Dewar in Austin, Texas. Upon joining the staff at Imperial College in 1977, his researches became focussed on computational mechanistic chemistry, NMR and chiroptical spectroscopies and Internet-based Chemical informatics, for which he was awarded the 2012 ACS Skolnik award. 0000-0002-8635-8390
0000-0002-8635-8390
[email protected]







